Researchers worldwide have been working to create a cell atlas of the human organism using single-cell sequencing for about five years. The method can be used to distinguish cell types because different genes are read - or expressed - in them. And this is how it works:
Letters from the nucleus
All cells of an organism carry the same genetic information in the cell nucleus. But why do skin cells have entirely different properties than heart muscle cells? Each cell produces only those gene copies that it needs for its function. The gene copies are called messenger RNA and carry the blueprint for a protein. You can think of messenger RNA as letters with building instructions that the nucleus sends to the cell's protein factories, the ribosomes.
Different cell types require different proteins - therefore, depending on the cell type, different letters with building instructions buzz around in the cells - in other words, each cell type has a characteristic composition of messenger RNA molecules. If these molecules are analyzed cell by cell, a complex bioinformatics procedure provides an exact idea of the individual cells that comprise tissues and organs. In 2020, the "Cell Atlas of the Adult Heart" was published in Nature.
Telltale patterns
If healthy cells from different tissues differ in expression patterns, this should also be the case for healthy and diseased cells from the same tissue, the Frankfurt researchers reasoned. They, therefore, examined the cardiac septum of patients who had received a new aortic valve. A leaky aortic valve overloads the heart muscle, causing the heart cells to enlarge in an unhealthy way; they are said to be hypertrophy. The researchers obtained biopsies of hypertrophic septum directly from the operating room of the heart surgery department at Frankfurt University Hospital.
Impaired communication makes heart cells sick
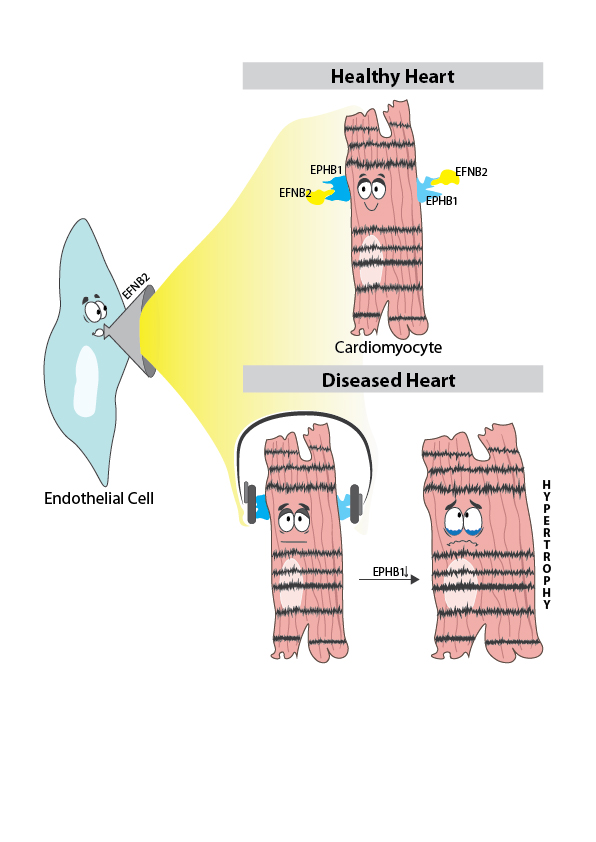
Using single-cell sequencing, they first assigned the cells to the cell types typical of heart tissue, such as cardiac muscle cells, connective tissue cells, endothelial cells (they line the blood vessels), and immune cells. They then determined the expression of additional genes for each cell type and compared these with a data set from healthy heart tissue that scientists from the Wellcome Sanger Institute had published in Nature."We noticed cardiac muscle cells in the diseased heart communicate significantly less with other cell types. In particular, we became aware of the gene for the receptor EPHB1, because it was only very lowly expressed in the diseased heart cells, meaning it had virtually stopped working," said the first author of the study Simone-Franziska Glaser from the Institute for Cardiovascular Regeneration at the Johann Wolfgang Goethe University Hospital. According to scientific literature, EPHB1 exchanges signals with a ligand called EFNB2. The latter is mainly located on endothelial cells. The researchers' hypothesis: the heart muscle cells no longer "hear" what the endothelial cells "have to say" and that's not good for them.The researchers confirmed their hypothesis in cell models: If the EPHB1 signaling pathway was artificially disrupted because the endothelial cells could not produce EFNB2, this stressed the cardiac muscle cells. They beat (contracted) more slowly and enlarged pathologically. If the heart muscle cells had contact with sufficient EFNB2, the cells remained normal-sized and beat at the correct pace."We are pleased to have been able to describe this mechanism for the first time," Glaser says. Her team plans to analyze the sequence data more deeply, including other parts of the heart and different severities of cardiac hypertrophy. "In this way, we will learn to understand how myocardial enlargement develops and what causes it to worsen - to develop a therapy for the diseased heart," she says.
Puplication: Nicin, L., Schroeter, S.M., Glaser, S.F. et al. A human cell atlas of the pressure-induced hypertrophic heart. Nat Cardiovasc Res 1, 174–185 (2022). https://doi.org/10.1038/s44161-022-00019-7
Scientific contact person: Dr. Simone-Franziska Glaser, Klinikum der Johann Wolfgang Goethe-Universität, Institut für kardiovaskuläre Regeneration, glaser(at)med.uni-frankfurt.de
Contact: Christine Vollgraf, Presse- und Öffentlichkeitsarbeit, Deutsches Zentrum für Herz-Kreislauf-Forschung (DZHK), Tel.: 030 3465 529 02, presse(at)dzhk.de