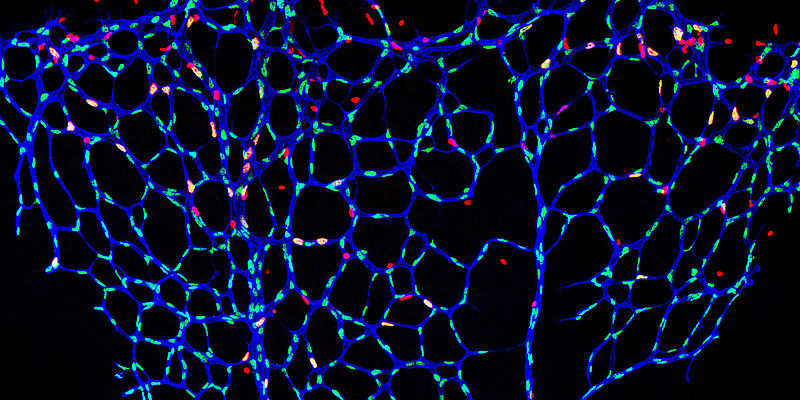
A complex network of blood vessels distributes blood to all organs of the body, ensuring that the body's cells are supplied with sufficient oxygen and nutrients to maintain heartbeat and brain function, for example. Blockages in blood vessels and thus impaired oxygen supply can lead to neuronal or cardiac cell death and culminate in a stroke or heart attack. Revascularisation - i.e. the restoration of blood supply and tissue regeneration - requires functional blood vessels. How this revascularisation of organs actually takes place and how it can be promoted is still an unsolved clinical problem.
As each organ fulfils a unique physiological function, the vascular branching patterns, i.e. the blood vessels, differ between the organs. How organ-specific vascular structures develop has long been a mystery.
From a therapeutic perspective, it is believed that a fundamental understanding of the organ-specific molecular control of vascular growth and branching provides a key to the development of personalised medicine strategies to combat cardiovascular and neurodegenerative diseases.
Pioneer cells move within the vessel walls
Scientists led by Professor Ferdinand le Noble, head of the Department of Cell and Developmental Biology at the Zoological Institute of the KIT, have now discovered that the activation of a new vascular cell type is a crucial element of organ-dependent variability in vascular branching. They call this cell type the endothelial L-spike cell or pioneer cell. Pioneer cells are located within the inner layer that lines the blood vessel, the so-called endothelium.
Using state-of-the-art imaging techniques, the researchers discovered that pioneer cells move within the vessel wall. As soon as they come into contact with specific signals produced by the local environment, these pioneer cells begin to form new blood vessels. In order to clarify the molecular identity of these signals, to identify the signal-producing cells and, above all, the mechanism by which these signals are perceived to promote the differentiation of pioneer cells, the scientists used a recently developed technique called single-cell sequencing.
Molecular cocktail encodes the time and place of blood vessel formation
"Single-cell sequencing combines detailed RNA sequencing of individual cells with bioinformatic analyses and enables precise identification of cell subtypes and the molecules that these cells synthesise for cell-cell communication," explains Dr Laetitia Préau, first author of the study. "Using this technique, we discovered that vascular patterning is encoded by a specific set of molecules that can only be sensed by a subset of endothelial cells to promote vascular growth."
This cocktail of molecules is organ-specific and encodes the instructions for how a new blood vessel can be formed at that particular place and time. The only recipients of this molecular code are the future pioneer cells, which decipher the code and initiate vascular growth processes.
Foundations for new therapeutic approaches
It turned out that several of these organ-specific vascular growth code molecules can potentially be manipulated pharmacologically, i.e. react to chemicals added from outside. Professor Ferdinand le Noble explains: "In order to explore the therapeutic possibilities, we are working together with experts from chemistry, tissue engineering and AI in our 3R Centre 3ROCKIT at the KIT Centre for Health Technologies (KITHealthTech). We hope to identify novel smart molecules to influence the vascular growth process, which could benefit patients with ischaemic cardiovascular diseases such as heart attacks and strokes, as well as certain forms of cancer."
The study was funded by the German Research Foundation (DFG) and carried out by KIT in collaboration with the German Center for Cardiovascular Research (DZHK) at its partner sites in Heidelberg and Munich and the Max Planck Institute for Molecular Biomedicine in Münster.
Link to the original publication: Parenchymal cues define Vegfa-driven venous angiogenesis by activating a sprouting competent venous endothelial subtype (Préau et al., 2024)
Source: press release KIT